The lab focuses on integrating multiple techniques to examine how genes are regulated during brain development and in neurodevelopmental disorders. We use in vivo rodent models, human tissues and in vitro cell culture to examine how genetic and environmental risk factors for neurodevelopmental disorders alter the epigenome, cellular interactions and animal behaviour.

Open Science Tools for Examining Microglia
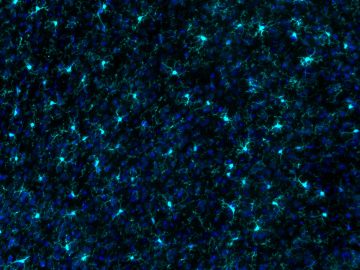
Microglia are a unique and highly plastic brain cell. The Ciernia lab has focused on developing new tools for analyzing microglial gene expression and function. We developed a web based R-Shiny application that allows users to compare their own gene lists against a curated set of microglial and brain disorder specific gene lists (Jao & Ciernia, 2021). The resulting MGEnrichment tool is available at
https://ciernialab.shinyapps.io/MGEnrichmentApp/ and further online documentation and datasets can be found at https://github.com/ciernialab/MGEnrichmentApp. We also have developed a novel method for microglial isolation and quantification of global levels of transcriptional regulators and histone modifications (Towriss et al, 2023). Our recent efforts have focused on open-source tools for analyzing microglial morphology and are currently under review (Kim, Pavlidis, & Ciernia, 2024). The analysis package and example images are all open source: https://github.com/ciernialab. Together these tools benefit the wider neuroscience community and specifically researchers interested in neuro-immune interactions in the brain.
Epigenomic Regulation in Microglia
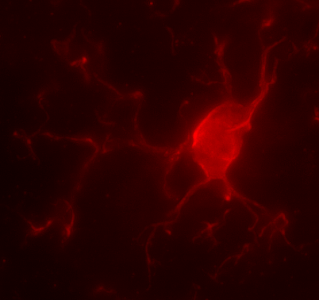
Microglia, the brain’s immune cells, directly interact with neurons during brain development to shape functional synaptic circuits. Dr. Ciernia profiled microglia, the brain’s immune cell population, in mice subjected to maternal immune activation in utero. Specifically, she examined the impact of maternal allergic asthma on microglial DNA methylation and gene expression in juvenile, female offspring. This was the first whole genome DNA methylation profiling from microglia and revealed distinct cell type specific regions of DNA methylation that appear critical for microglial function (Vogel Ciernia, et al., 2018). Dr. Ciernia has since expanded these findings to elucidate how chromatin structure interacts with genetic variation to regulate immune gene expression (Ciernia et al., 2020; Kim & Ciernia, 2020) and how microglial gene expression is impacted by sex (Sullivan & Ciernia, 2022). Recent work from the Ciernia lab uncovered novel histone regulation pathway in microglial inflammation (Meleady et al., 2023). We specifically found that Hdac3 acts as a negative regulator of histone acetylation at pro-inflammatory genes in microglia, and that Hdac3 inhibition enhances neuroprotective functions of microglia in response to LPS through reduced nitric oxide release and increased phagocytosis (reviewed Rosete & Ciernia, 2024). We recently also identified unique metabolic pathways underling different forms of microglial immune memory, leading to the discovery that shifts in aerobic glycolysis are a critical step in the induction of innate immune tolerance in vitro (Towriss, MacVicar, and Ciernia, 2023) and in vivo (Kim et al., 2024). Together, our work is uncovering novel epigenetic regulatory mechanisms governing microglial control of brain inflammation.
Check out a recent talk by Dr. Annie Ciernia on this project.
Gut-Microbiome-Brain Microglia Axis Regulation in Development and Disease
In the healthy brain, the gut microbiome releases metabolites that signal to microglia, the brain’s innate immune cells. In the context of gut inflammation and microbiota disruption as in Inflammatory bowel disease (IBD), gut-brain communication may be impaired. We hypothesized thatin altered gut microbe composition in IBD leads to dysfunction in the brain and behaviour. Consistent with this hypothesis, IBD patients experience higher rates of depression, anxiety, and social impairments, with childhood onset often resulting in long-lasting impacts. The Ciernia lab in collaboration with Dr. Carolina Tropini developed a novel pediatric model of IBD where gut inflammation drives altered sexual de velopment and behaviour in males (Sullivan, et al., 2024 and this commentary). We found that disruptions to the gut microbiota converge to influence brain function and behaviour including impacts on hormone and metabolite signalling to microglia. We have also recently expanded our efforts to develop a new model in which human patient or health control microbiota are transplanted into germ free mice lacking their own endogenous microbes. The resulting microbiome humanized-mice are then be used to study how alterations in the human microbiota in patient populations impacts the gut-brain axis, behaviour and disease risk.
velopment and behaviour in males (Sullivan, et al., 2024 and this commentary). We found that disruptions to the gut microbiota converge to influence brain function and behaviour including impacts on hormone and metabolite signalling to microglia. We have also recently expanded our efforts to develop a new model in which human patient or health control microbiota are transplanted into germ free mice lacking their own endogenous microbes. The resulting microbiome humanized-mice are then be used to study how alterations in the human microbiota in patient populations impacts the gut-brain axis, behaviour and disease risk.
Chromatin Remodeling in Neuronal Development and Neurodevelopmental Disorders
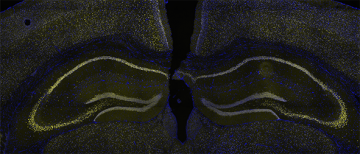
Mutations in 12 of the 29 subunit genes that compose the BAF (Brg1 Associated Factor) chromatin remodelling complex have been identified in ASD and Intellectual Disability (ID), making it one of the most common genetically implicated protein complexes in neurodevelopmental disorders. However, it is unclear how mutations in this epigenetic regulator lead to childhood disorders. My lab is examining Baf53b, a subunit that confers neuronal specificity to the neuronal version of the BAF complex (nBAF). Baf53b is expressed exclusively in neurons and is only found within the nBAF complex, making it an ideal target for genetic manipulations to elucidate nBAF’s role in brain development.I previously showed that heterozygous loss of Baf53b impaired dendritic spine development, synaptic plasticity, and long-term memory in the adult mouse (Vogel Ciernia, et al., 2013). These deficits were reversible by manipulating structural proteins at the neuronal synapse (Vogel Ciernia et al. 2017), suggesting a critical link between chromatin remodelling impairments and synaptic deficits, and new potential therapeutic avenues for brain disorders. My lab will test the hypothesis that conditional deletion of Baf53b within specific neuronal populations during brain development in mice disrupts chromatin regulation. We will link the gene regulatory functions of nBAF to the development of relevant behaviours and deficits in neuronal connectivity and synapse maturation, allowing us to pinpoint the cell types, time points and gene targets regulated by nBAF. Our findings will provide mechanistic insight into a unique, neuronal epigenetic mechanism that is critical for normal brain development and highly relevant to therapeutic development for numerous childhood disorders including ASD, ID and epilepsy. Check out a recent talk by Dr. Megan Rowland from the Ciernia lab on this project.