The lab focuses on integrating multiple techniques to examine how genes are regulated during brain development and in neurodevelopmental disorders. We use in vivo rodent models, human tissues and in vitro cell culture to examine how genetic and environmental risk factors for neurodevelopmental disorders alter the epigenome, cellular interactions and animal behavior.
Signatures of Microglial Dysfunction across Autism Spectrum Disorders (ASD) Models
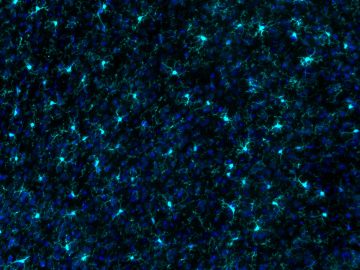 Gene expression patterns and cellular morphology changes in human post-mortem ASD brain indicate increased microglial activation. However, it is unclear how changes in microglial function contribute to ASD, and if the observed immune signatures are a cause or consequence of altered brain development. To parse the role of microglia in ASD, are examining both genetic and environmental mouse models to address: When do microglial abnormalities arise during brain development? We hypothesize that the microglial transcriptome is altered in ASD mouse models across development and at least a subset of microglial genes are commonly disrupted across models, resulting in shared pathways of microglial dysfunction and altered microglial morphology. To test this hypothesis, we are examining changes in microglial gene expression and morphology from genetic and environmental ASD risk mouse models at multiple developmental timepoints. Gene expression changes will then be incorporated into network-level bioinformatics analyses to identify and test novel gene targets and epigenomic regulatory pathways dysregulated across time in microglia from ASD. We are also addressing the question of How do altered microglial-neuronal interactions impact cellular development? We hypothesize that interactions between neurons and microglia lead to the neuroinflammatory phenotype found in human ASD brain. To test this hypothesis, we are examining how mutant microglia from different ASD models impact wildtype neuronal development. The long-term goal of this work is to identify common pathways of dysfunction that are conserved across different mouse models and in human ASD that can serve as novel biomarkers or therapeutic targets.
Gene expression patterns and cellular morphology changes in human post-mortem ASD brain indicate increased microglial activation. However, it is unclear how changes in microglial function contribute to ASD, and if the observed immune signatures are a cause or consequence of altered brain development. To parse the role of microglia in ASD, are examining both genetic and environmental mouse models to address: When do microglial abnormalities arise during brain development? We hypothesize that the microglial transcriptome is altered in ASD mouse models across development and at least a subset of microglial genes are commonly disrupted across models, resulting in shared pathways of microglial dysfunction and altered microglial morphology. To test this hypothesis, we are examining changes in microglial gene expression and morphology from genetic and environmental ASD risk mouse models at multiple developmental timepoints. Gene expression changes will then be incorporated into network-level bioinformatics analyses to identify and test novel gene targets and epigenomic regulatory pathways dysregulated across time in microglia from ASD. We are also addressing the question of How do altered microglial-neuronal interactions impact cellular development? We hypothesize that interactions between neurons and microglia lead to the neuroinflammatory phenotype found in human ASD brain. To test this hypothesis, we are examining how mutant microglia from different ASD models impact wildtype neuronal development. The long-term goal of this work is to identify common pathways of dysfunction that are conserved across different mouse models and in human ASD that can serve as novel biomarkers or therapeutic targets.
Epigenomic Regulation of Microglial Development
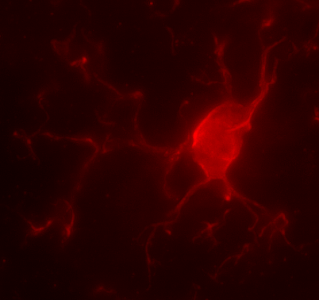
The goal of this project is to characterize and causally test the interplay of multiple epigenomic regulatory mechanisms in coordinating microglial maturation during normal brain development. During maturation microglia must coordinate specific patterns of gene expression for migration, differentiation, and establishment of a homeostatic state in the brain. This precise regulation of gene expression requires interactions with the maturing nervous system and is orchestrated by epigenetic mechanisms that allow both dynamic control of gene expression and stable, long-term cellular differentiation. Within the brain microglia self-renew for the lifetime of the animal, suggesting that that epigenetic processes dictating early microglial development may have long-lasting impacts on cellular function.Previous work has identified key developmental enhancers that are epigenetically regulated during microglial differentiation. Our work is expanding on this knowledge to identify regulatory regions that are dynamically controlled during microglial maturation and how they form a final, stable homeostatic state in the adult brain. We are utilizing a variety of epigenomic sequencing techniques to answer this question including CUT&Tag-Seq, ATAC-Seq and RNA-Seq. Using advanced bioinformatic and statistical modelling we can combine the epigenomic information to understand microglial gene regulation across development. Check out a recent talk by Dr. Annie Ciernia on this project.
Chromatin Remodeling in Neuronal Development and Neurodevelopmental Disorders
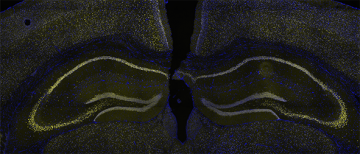
Mutations in 12 of the 29 subunit genes that compose the BAF (Brg1 Associated Factor) chromatin remodelling complex have been identified in ASD and Intellectual Disability (ID), making it one of the most common genetically implicated protein complexes in neurodevelopmental disorders. However, it is unclear how mutations in this epigenetic regulator lead to childhood disorders. My lab is examining Baf53b, a subunit that confers neuronal specificity to the neuronal version of the BAF complex (nBAF). Baf53b is expressed exclusively in neurons and is only found within the nBAF complex, making it an ideal target for genetic manipulations to elucidate nBAF’s role in brain development.I previously showed that heterozygous loss of Baf53b impaired dendritic spine development, synaptic plasticity, and long-term memory in the adult mouse (Vogel Ciernia, et al., 2013). These deficits were reversible by manipulating structural proteins at the neuronal synapse (Vogel Ciernia et al. 2017), suggesting a critical link between chromatin remodelling impairments and synaptic deficits, and new potential therapeutic avenues for brain disorders. My lab will test the hypothesis that conditional deletion of Baf53b within specific neuronal populations during brain development in mice disrupts chromatin regulation. We will link the gene regulatory functions of nBAF to the development of relevant behaviours and deficits in neuronal connectivity and synapse maturation, allowing us to pinpoint the cell types, time points and gene targets regulated by nBAF. Our findings will provide mechanistic insight into a unique, neuronal epigenetic mechanism that is critical for normal brain development and highly relevant to therapeutic development for numerous childhood disorders including ASD, ID and epilepsy. Check out a recent talk by Dr. Megan Rowland from the Ciernia lab on this project.